Research
The genomic architecture of adaptive radiation
Adaptive radiation—the evolutionary phenomenon where a single ancestral lineage diversifies into a variety of adaptive forms—has fascinated evolutionary biologists since Darwin. These extraordinary morphological and ecological diversifications have long captivated biologists, and example radiations have been model systems to study the link between ecology and speciation. Adaptive radiations in remote settings such as islands are especially intriguing and even paradoxical. How can rapid species diversification occur from a genetically bottlenecked founder population?
Our research addresses this question by studying the Hawaiian Metrosideros and understand the evolutionary basis of island adaptive radiation in plants. The Metrosideros group is one of the most widely dispersed flowering plants in the Pacific Basin, growing from the Philippines to Chile. The Hawaiian archipelago is particularly rich with 23 morphotypes that are non-randomly distributed across the island. Through collaboration with Elizabeth Stacy (UNLV), our research implements evolutionary and population genomic approaches to study the biological basis of the Metrosideros adaptive radiation
Related work: Choi et al. 2021. PNAS; Choi et al. 2020. Mol Biol Evol
Evolution meeting 2022 talk on Hawaiian Metrosideros genomics: link
Telomere variation and life history evolution
Structural variations (SVs) are large complicated sequence changes across the genome. Its evolutionary significance is largely elusive due to the difficulties of its detection. Our research focuses on detecting and studying the evolutionary consequence of SVs, by applying state-of-the-art genome sequencing technology and collecting phenotypic data from the field and the lab.
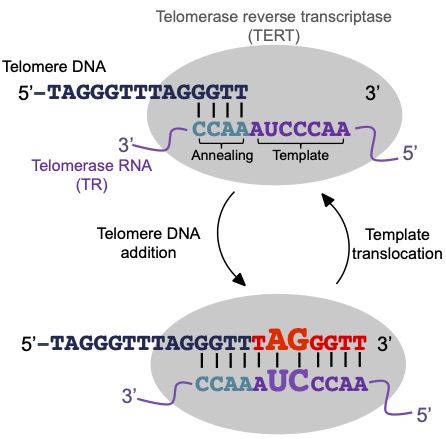
One particular SV within the chromosome are those found in the telomere. Telomeres maintain chromosome ends from damage and are under functional constraint as aberrations can have dire consequences. Paradoxically, telomere lengths vary naturally between individuals, for instance telomere lengths can vary over 10-fold in Arabidopsis thaliana. But why this variability is observed in plants is not known.
Our previous research has discovered plant telomeres are a target of selection and confers adaptive benefits in specific life history or ecological conditions. Importantly the study suggests telomeres are novel genetic source of adaptive traits in plants, emphasizing chromosome structures as understudied genetic variations of plant ecological diversification. Our research aims to understand the molecular link between flowering and telomere structure in plants, to understand the biological basis of chromosome structure and life history trait evolution.
Related work: Kumawat et al. 2023. bioRxiv; Choi et al. 2021. The Plant Cell
TAGC 2024 talk on Mimulus telomere evolution: link
Developing and applying genomic techniques for evolutionary research
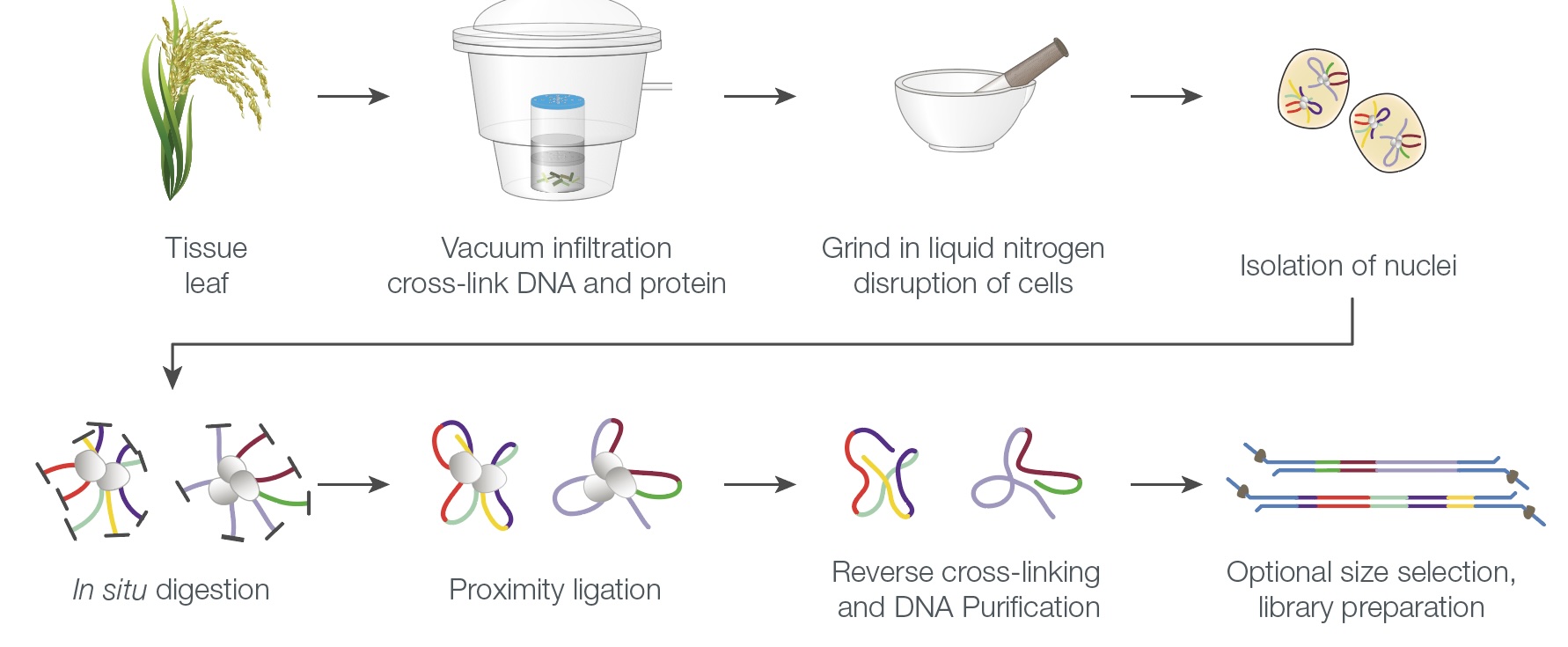
Long read sequencing is revolutionary for evolutionary biology research. Generating high quality genomic resources is a reachable goal for non-model organisms. Our research utilizes state-of-the-art genomic sequencing technologies (in particular nanopore sequencing) to generate high-quality reference genome as a basis for studying the genome-wide variations of our plants of interest. We are also interested in using nanopore sequencing to develop and implement techniques that takes a deeper inference of the genomic variation. One such example is PoreC sequencing which is a long read sequencing based chromosome contact assay. Finally we are focused on developing bioinformatic methods to use the long read sequencing data or other genome-wide variation data to study the evolutionary basis of adaptation and speciation.
Related work: Choi et al. 2022. Biorxiv; Choi et al. 2021. PNAS; Choi et al. 2020. Genome Biol